Introduction
Prostate cancer (PCa) is the second most commonly diagnosed cancer globally and the fifth leading cause of cancer-related deaths among men. In 48 countries, PCa is the leading cause of mortality among male cancer patients1. Given the global aging population, it is expected that the burden of PCa will continue to increase. Moreover, advanced age is not the only factor contributing to the growing problem. Studies conducted in various countries have identified new risk factors for PCa, including obesity, diabetes mellitus2 3, dietary habits, and vitamin E supplements4.
As a result, the development of diagnostic and treatment methods for PCa is an urgent issue that requires appropriate action. Basic PCa diagnostics include digital rectal examination (DRE), serum prostate-specific antigen (PSA) measurement, and transrectal ultrasound (TRUS). PSA screening has contributed to a more than 50% reduction in prostate cancer mortality5; however, it has also led to the significant issue of overdiagnosis and overtreatment of non-aggressive PCa6. Consequently, the focus has shifted toward preferential diagnosis and treatment of aggressive PCa.
Currently, the primary method for diagnosing prostate cancer is histomorphological evaluation of prostate biopsies7 8. A key component of morphological assessment is tumor differentiation using the Gleason grading system, which is based on the architectural models of tumor growth. The Gleason score is also used for subsequent ISUP (International Society of Urological Pathologists) classification9 10, which provides more accurate stratification of prostate tumors and helps reduce the frequency of active radical treatments in patients with clinically insignificant PCa.
Tumor Malignancy Assessment
The Gleason score is a method for evaluating the malignancy of cancerous tumors and predicting their aggressiveness. Developed by Donald Gleason in 1966, this system is based on the histological evaluation of the architecture of tumor tissues11. The primary principle of the Gleason score involves classifying tumor glands according to their level of differentiation: from well-differentiated, resembling normal tissue, to poorly differentiated, which significantly differs from normal cells. In this method, the pathologist selects the two most prevalent histological patterns of cellular structure in the sample and assigns each a score from 1 to 5. Each score represents a specific cellular structure as follows12:
1. The gland cells are small and well-defined;
2. The gland cells are spaced far apart;
3. The gland cells have an irregular shape;
4. Few cells have a regular shape, forming a neoplastic mass;
5. Absence of glandular cells.
The sum of these two scores forms the so-called 'Gleason score.' According to the decision of the International Society of Urological Pathologists (ISUP, 2014 conference), which was included in the WHO recommendations (2016), Gleason score calculations for prostate cancer diagnostics should utilize grades 3-5. The first two grades were excluded10 13 for several reasons. First, scores of 4 are extremely rare in surgical materials. Second, in the modern interpretation, the first-grade pattern corresponds to benign hyperplasia and should not be classified as adenocarcinoma. Third, the second-grade pattern is considered a secondary manifestation of the third-grade pattern, rather than an independent pathology. As a result, the Gleason score can range from 6 to 10. A higher score indicates a more aggressive and less differentiated tumor, correlating with a poorer prognosis for the patient. The current version of the Gleason score not only describes the degree of cellular deviation of the cancerous tumor from healthy glandular epithelium but also categorizes patients into five prognostic groups based on expected five-year recurrence-free survival. For instance, group one has a total Gleason score of 6 (3+3) with a survival probability of 97%, while group five has a total score of 9–10 with a survival probability of 49%.
Despite the high level of algorithmization in tumor classification using the Gleason score, significant fluctuations in specialist conclusions are sometimes observed14 15 16. This is largely due to the considerable variety of histological structures, even within the same tumor. Variability in assessment also depends on errors in tissue sampling by urologists and the limited amount of material. The qualifications and even the personality traits of the specialist, which can lead to either optimistic or pessimistic prognoses, also significantly influence the results. Therefore, prostate cancer diagnostics, including the use of the Gleason score, requires the development of new reliable and reproducible methods that minimize the variability in conclusions.
Improvement of the Tumor Pathohistological Assessment
As mentioned above, the Gleason scoring system underwent significant changes in the 2000s12. In addition to increasing the number of biopsy sites, the original assessment templates were revised and expanded. New methods for processing biopsy material were introduced to improve the discrimination between different tissue types. Additionally, it was suggested to account for the ratio of tumor to normal tissue in biopsy samples, along with other parameters, to enhance diagnostic and prognostic accuracy.
One of the most promising approaches to addressing these issues has been the application of artificial intelligence (AI). Research in this area is actively being conducted by centers in various countries17 18 19 20. In particular, deep learning neural networks are being used for the automatic identification and grading of Gleason patterns with promising results. Modern machine learning algorithms and convolutional neural networks (CNNs) allow for the standardization of the diagnostic process, improving objectivity, accuracy, and reproducibility.
Aim
The aim of this work was to develop and implement a neural network-based algorithm for detecting Gleason patterns in whole-slide histological images.
Application of Neural Network Models
CNNs are a key tool in solving practical tasks in the field of computer vision, as the convolution operator allows for the sequential extraction and processing of features at various levels: from basic (lines, points) to complex (object contours). In digital form, these features represent multidimensional vectors used for image classification. These accumulated features can be transformed into segmented areas of the original image through reverse transformation. This approach, using neural networks for classification and segmentation, is known as the Encoder-Decoder architecture. In general, models of this architecture consist of two parts (Figure 1).
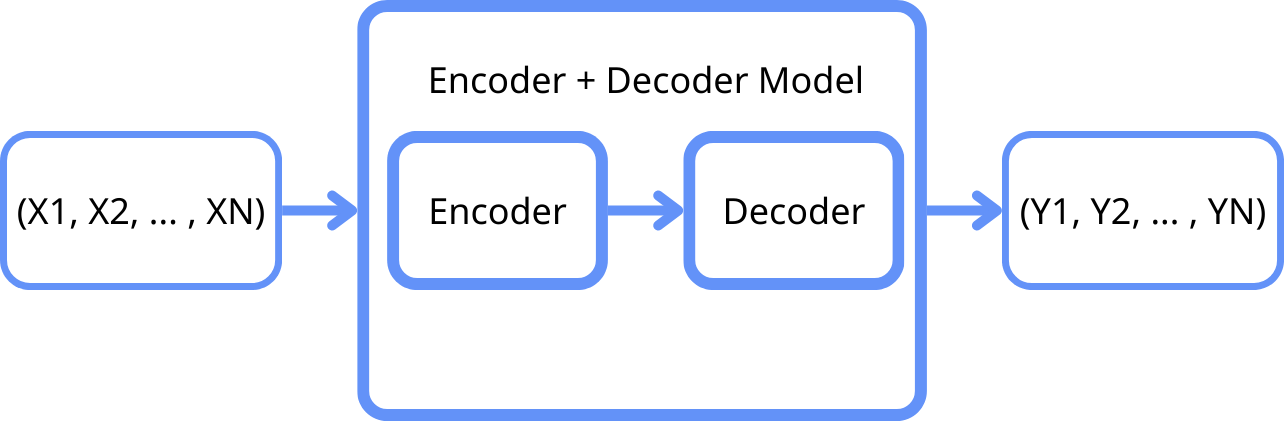
Figure 1: General scheme of the model architecture.
The Encoder performs the function of reducing the dimensionality of input data by extracting the most significant features. In the context of histological images, the encoder typically consists of several convolutional layers that sequentially process the input images, extracting high-level features. These features include the textural and architectural characteristics of tissues, which are crucial for differentiating between malignant and normal cells.
After the encoding stage, the compressed data representation is passed to the Decoder, which consists of several layers that perform the reverse of the convolution, restoring the data to its original resolution or to a format required for further analysis. During the decoding process, the neural network learns to reconstruct the features and structure of the original images.
In the context of prostate cancer diagnostics, such models can be used to analyze images obtained from biopsy samples. Histological images are inherently complex, with a high level of intra-class variability, making their analysis labor-intensive for specialists. Models of this architecture can improve automation of the analysis due to their ability to account for spatial and structural dependencies in the data. The primary representative of this family of models is U-Net.
U-Net and SegFormer Models for Histological Image Segmentation
U-Net is one of the most popular architectures for image segmentation tasks, including medical imaging, due to its ability to efficiently capture both local and global contexts. The core of U-Net is a symmetrical structure consisting of an encoder and decoder, which allows the model to restore spatial information at various resolution levels. U-Net demonstrates high accuracy and stability when working with limited datasets, making it a suitable choice for the initial stages of research.
However, with the emergence of transformers, which initially demonstrated excellent results in text-processing tasks, their potential has also been successfully adapted for image analysis. The SegFormer model21 combines the advantages of transformers and multilayer perceptrons (MLPs), enabling it to capture long-range dependencies and the global context of images. This is particularly useful for histological images, where it is important to consider the relationships between different regions. SegFormer's hierarchical encoder structure also allows the model to handle variations in resolution and object scales, making it flexible and adaptable for image segmentation.
Moreover, SegFormer uses a simple MLP-based decoder that effectively aggregates information from different levels, combining local and global attention. This simplifies the architecture and reduces computational costs, which is a significant advantage when working with large volumes of data. Several studies have shown that SegFormer outperforms U-Net in segmentation accuracy across various datasets, including medical images22.
The use of SegFormer, in combination with different encoders such as EdgeNeXt or ResNet, has improved the accuracy of cancer segmentation in histological images, significantly reduced the number of model parameters, and increased processing speed compared to U-Net-based architectures.
Challenges in Analyzing Prostate Histological Images
The analysis of histological images using AI has been previously discussed in the literature, highlighting the following key issues and challenges23:
1. Lack of Data: One of the main problems in the field of medical image analysis is access to high-quality and diverse datasets. Successful training of neural networks requires a large volume of annotated data, which can be quite limited in the medical field.
2. Data Heterogeneity: Histological images can vary significantly depending on the equipment used, sample preparation methods, and evaluation criteria. Even within the same literature source, there may be articles with different classifications of tissue patterns24 25. These polymorphisms and other factors can create challenges in training universal models.
3. Data Size: Medical histological images tend to be very large. It is not uncommon to encounter images larger than 50,000 x 50,000 pixels, while deep neural network training typically uses lower resolutions due to limited computational resources. Splitting images into patches—dividing them into smaller fragments—can help address this issue, but it may lead to a loss of accuracy or even data loss. Nevertheless, models trained on patches are generally preferred over those trained on full images26.
Thus, when choosing a neural network architecture, it is important to consider all of the aforementioned challenges to achieve the highest possible accuracy.
Material and Methods
Tested Variants of Neural Network Architectures
In this study, a series of experiments were conducted to improve the neural network architecture for analyzing PCa histological images. The research object consisted of whole-slide histological images27, WSI scans of prostate cancer patients in *.svs format with 40x magnification and an average spatial resolution of 0.258 microns per pixel. The biopsy material, stained with hematoxylin and eosin, included both healthy samples and samples with pathological areas of varying Gleason score malignancy. The full dataset comprised 113 scans, annotated by three expert pathologists. Due to the very high resolution of the WSI scans, a tiled segmentation method was applied28. This approach allowed for efficient processing of large data volumes by dividing the scans into smaller fragments to improve the quality and accuracy of neural network training. ImageNet standardization was applied to each tile.
Various architecture combinations were tested, with encoders including VGG16, ResNetV2_50x1_bit29, Mit-b0, and EdgeNeXt-small30, and decoders such as U-Net31 32 and SegFormer. The models were developed using the PyTorch and timm libraries.
Since the number of tiles containing cancerous cells was significantly smaller than those with healthy cells, there was a strong class imbalance. To address this during neural network training, a specific ratio of tiles with and without cancer was used in each batch.
To increase the likelihood of including challenging non-cancerous tiles (where the model often made mistakes) in the training set, a "definitely not cancer" class was added. Tiles containing this class were prioritized during training. However, this approach did not improve model metrics, so the "definitely not cancer" class was not used in further training.
Additionally, earlier versions of the trained models frequently misclassified healthy areas with seminal vesicles and nerve bundles as cancerous. As a result, the corresponding classes, "seminal vesicles" and "nerve bundles," were added to the training data. However, the increased frequency of tiles with these classes did not lead to significant metric improvement. Nonetheless, the labeling and inclusion of these classes should be considered in future model training, as novice specialists frequently misclassify these areas.
The algorithm for determining tissue in the original images, which initially incorporated brightness thresholding of the image channels' dispersion, was improved by using a more complex saturation channel threshold in the HSV color space, followed by Gaussian blurring and hole filling.
To improve classification accuracy, an optimal binary mask threshold was calculated on validation data, ensuring balanced values between precision and recall.
Additionally, experiments were conducted with dynamically cropping tiles from the original *.svs images, as well as cropping mask tiles from full images in *.png format. However, tests showed that this approach was slower, so the decision was made to load pre-cropped tiles from disk.
To ensure high accuracy in detecting pathological areas at the periphery of the tile images, a segmentation process with four steps of tile shifts was implemented: 1) no shift, 2) half the tile width shift, 3) half the tile height shift, and 4) diagonal shift. Each shift, with weighted masks where the central areas of the tiles had the maximum weight, decreasing toward the periphery, was then integrated into a single mask. This approach provides high segmentation accuracy across the entire image and prevents the appearance of artifacts at the tile borders, which are typical in the single-pass method.
Further testing of the models on whole-slide images with four passes showed that increasing the input tile size from 256x256 to 512x512 improved segmentation accuracy and increased the overall scan processing speed.
Comparison of Tested Neural Networks
The tested model architectures were compared across several parameters:
1. Number of parameters: This includes all the weights and biases used in the model. This metric influences the amount of memory consumed by the model.
2. F1-score: This is a combined metric that accounts for both precision and recall. It is used to assess the quality of binary classification, especially in cases where classes are imbalanced:
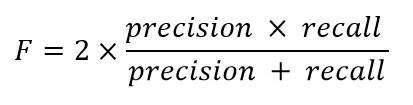
Where precision is the ratio of correctly classified positive examples to the total positive examples identified by the model, and recall is the ratio of correctly classified positive examples to the total true positive examples in the data.
3. Batches per second (validation): This refers to the number of data samples the model can process per second. This metric is calculated during the network's application to the validation set and reflects the network's efficiency and performance.
Table 1 presents a summary of the comparison of the tested models.
|
Experiment Group |
Model Name |
Number of Parameters (M) |
F1 Score on Tiles |
Batches per Second |
|
256x256, class 3 |
|
|
|
|
|
|
unet-vgg16_bn |
18,60 |
0,820 |
5,6 |
|
|
unet-edgenext_small |
7,660 |
0,835 |
16,2 |
|
|
unet-resnetv2_50x1_bit |
13,60 |
0,831 |
9,2 |
|
|
segformer-mit_b0 |
3,360 |
0,833 |
41,4 |
|
|
segformer-edgenext_small |
5,330 |
0,837 |
31,3 |
|
|
segformer-resnetv2_50x1_bit |
8,670 |
0,835 |
22,8 |
|
512x512, class 3 |
|
|
|
|
|
|
segformer-mit_b0 |
3,360 |
0,828 |
9,3 |
|
|
segformer-edgenext_small |
5,330 |
0,835 |
8,6 |
|
|
segformer-resnetv2_50x1_bit |
8,670 |
0,834 |
6,3 |
|
512x512, class 4 |
|
|
|
|
|
|
segformer-mit_b0 |
3,360 |
0,745 |
9,3 |
|
|
segformer-edgenext_small |
5,330 |
0,769 |
8,6 |
|
|
segformer-resnetv2_50x1_bit |
8,670 |
0,748 |
6,3 |
Table 1: Results of the Tested Models
Results
To solve the segmentation tasks for Gleason grades of varying malignancy, two neural networks were trained using the SegFormer architecture, modified by integrating the EdgeNeXt model (small variant) as a backbone (encoder). The first neural network identifies tissue regions with combined Gleason patterns of grades 3, 4, and 5, while the second identifies regions with combined patterns of grades 4 and 5. This approach aimed to increase the number of cancerous training tiles and thus improve the results. Training was performed using tiles sized 512x512 pixels, extracted from WSI scans at a reduced magnification of 5x. Only tiles where biological tissue occupied more than 25% of the area were used for training. A data batch with a specific ratio of tiles was created: one tile with cancerous cells for every three tiles without. The total number of tiles used to train the networks was 15,522, with 94 WSI scans. A total of 19 WSI scans were used for the validation set.
The best accuracy was achieved with input image sizes of 512x512 (corresponding to the tile size) and a batch size of 8, resulting in an F-score of 0.835 and 0.769 on the validation set for models identifying Gleason patterns of grades 3, 4, 5, and 4, 5, respectively. The models were trained for 10 epochs using the AdamW optimizer and a loss function that combined Focal Loss and Tversky Loss (parameters alpha=0.25, beta=0.75). Training was performed on a GeForce GTX 1080 Ti GPU, with each model's training taking approximately one hour.
Figure 2 shows a fragment of a WSI scan comparing the results of the model identifying combined Gleason patterns of grades 3, 4, and 5 in multi-pass segmentation mode with the annotations made by specialists.
The average segmentation time for Gleason patterns 3 and 4 on an image with a resolution of 80,000x57,000, using the multi-pass mode, was approximately 58 seconds on a GeForce GTX 1080 Ti (about 112 seconds for an image with a resolution of 66,000x124,000). The fast single-pass segmentation mode processed images four times faster but resulted in lower accuracy at the tile borders.
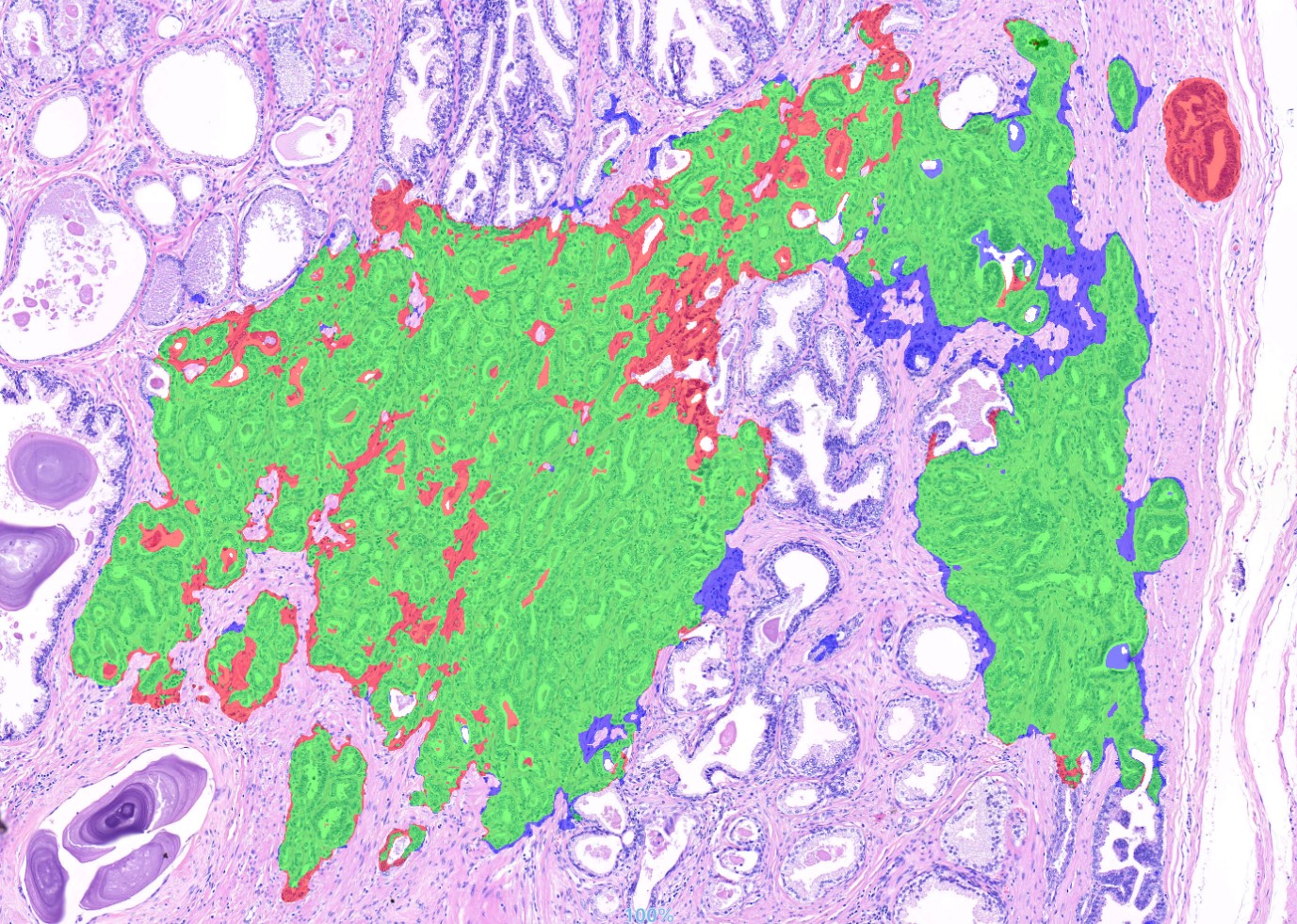
Figure 2: Comparison of the final model's results on a fragment of a WSI scan: green indicates areas where the annotations by specialists and the neural network matched, red indicates areas where the neural network highlighted a benign (unnecessary) region, and blue indicates areas where the neural network missed a malignant region.
Discussion
During testing at the Department of Pathological Anatomy of the Belarusian State Medical University, the neural network successfully detected almost all associated areas of pathological tissue, though it made some errors in determining the sizes of these areas during segmentation. In several cases, the network identified small, isolated lesions that were missed by the expert due to their size. Thus, the neural network demonstrated its potential effectiveness as a tool for preliminary research in screening mode, as well as an assistant in forming diagnostic conclusions. The model could be especially valuable in situations with limited access to qualified human specialists.
Conclusion
The use of artificial neural networks with the SegFormer architecture, supplemented by the EdgeNeXt (small variant) subnet, not only provided high repeatability of segmentation results but also significantly accelerated the diagnostic process, reducing the time for analyzing WSI scans to just a few minutes. Additionally, the use of deep learning improved the accuracy in identifying tumor regions and their malignancy grades. This enables more precise predictions of disease progression and better treatment planning.
Future research on the described neural network models aims to reduce errors in misidentifying seminal vesicles as cancerous areas and to expand the annotated dataset with more samples containing Gleason grade 5 in order to train a specialized model.
Conflict of Interest: Authors state that no conflict of interest exists.
References
1 Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209-249. doi:10.3322/caac.21660.
2 Tsilidis KK, Allen NE, Appleby PN, et al. Diabetes mellitus and risk of prostate cancer in the EuropeanProspectiveInvestigation into Cancer and Nutrition. Int. J. Cancer. 2015;136(2):372-381. doi:10.1002/ijc.28989.
3 Andersson SO, Wolk A, Bergstrom R, et al. Body size and prostate cancer: a 20-year follow-up study among 135006 Swedish construction workers. J Natl Cancer Inst. 1997;89(5):385-389. doi:10.1093/jnci/89.5.385.
4 Klein EA, Thompson IM, Tangen CM, et al. Vitamin E and the Risk of Prostate Cancer. JAMA. 2011;306(14):1549. doi:10.1001/jama.2011.1437.
5 Ilic D, Djulbegovic M, Jung JH, et al. Prostate cancer screening with prostate-specific antigen (PSA) test: a systematic review and meta-analysis. BMJ. 2018;362:k3519. doi:10.1136/bmj.k3519.
6 Eklund M, Jaderling F, Discacciati A, et al. MRI-Targeted or Standard Biopsy in Prostate Cancer Screening. N Engl J Med. 2021;385(10):908-920. doi:10.1056/NEJMoa2100852.
7 Descotes J-L. Diagnosis of prostate cancer. Asian J Urol. 2019;6(2):129-136. doi:10.1016/j.ajur.2018.11.007.
8 Ahdoot M, Wilbur AR, Reese SE, et al. MRI-Targeted, Systematic, and Combined Biopsy for Prostate Cancer Diagnosis. N Engl J Med. 2020;382(10):917-928. doi:10.1056/NEJMoa1910038.
9 Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. American Journal of Surgical Pathology. 2016;40(2):244-252. doi:10.1097/PAS.0000000000000530.
10 van Leenders GJLH, van der Kwast TH, Grignon DJ, et al. The 2019 International Society of Urological Pathology (ISUP) Consensus Conference on Grading of Prostatic Carcinoma. Am J Surg Pathol. 2020;44(8):e87-e99. doi:10.1097/PAS.0000000000001497.
11 Gleason DF, Mellinger GT. Prediction of Prognosis for Prostatic Adenocarcinoma by Combined Histological Grading and Clinical Staging. J Urol. 2017;197(2S):S134-S139. doi:10.1016/j.juro.2016.10.099.
12 Epstein JI, Allsbrook WC, Amin MB, Egevad LL. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. American Journal of Surgical Pathology. 2005;29(9):1228-1242. doi:10.1097/01.pas.0000173646.99337.b1.
13 Baydar DE, Epstein JI. Gleason grading system, modifications and additions to the original scheme. TJPATH. 2009;25(3):59. doi:10.5146/tjpath.2009.00975.
14 Dere Y, Celik OI, Celik SY, et al. A grading dilemma; Gleason scoring system: Are we sufficiently compatible? A multi center study. Indian J Pathol Microbiol. 2020;63(Supplement):S25-S29. doi:10.4103/IJPM.IJPM_288_18.
15 La Taille A de. Evaluation of the interobserver reproducibility of gleason grading of prostatic adenocarcinoma using tissue microarrays. Human Pathology. 2003;34(5):444-449. doi:10.1016/s0046-8177(03)00123-0.
16 Oyama T, Allsbrook WC, Kurokawa K, et al. A Comparison of Interobserver Reproducibility of Gleason Grading of Prostatic Carcinoma in Japan and the United States. Archives of Pathology & Laboratory Medicine. 2005;129(8):1004-1010. doi:10.5858/2005-129-1004-ACOIRO.
17 Soerensen SJC, Fan RE, Seetharaman A, et al. Deep Learning Improves Speed and Accuracy of Prostate Gland Segmentations on Magnetic Resonance Imaging for Targeted Biopsy. Journal of Urology. 2021;206(3):604-612. doi:10.1097/JU.0000000000001783.
18 Yamamoto Y, Tsuzuki T, Akatsuka J, et al. Automated acquisition of explainable knowledge from unannotated histopathology images. Nat Commun. 2019;10(1). doi:10.1038/s41467-019-13647-8.
19 Kott O, Linsley D, Amin A, et al. Development of a Deep Learning Algorithm for the Histopathologic Diagnosis and Gleason Grading of Prostate Cancer Biopsies: A Pilot Study. Eur Urol Focus. 2021;7(2):347-351. doi:10.1016/j.euf.2019.11.003.
20 Lucas M, Jansen I, Savci-Heijink CD, et al. Deep learning for automatic Gleason pattern classification for grade group determination of prostate biopsies. Virchows Arch. 2019;475(1):77-83. doi:10.1007/s00428-019-02577-x.
21 Xie E, Wang W, Yu Z, Anandkumar A, Alvarez JM, Luo P. SegFormer: Simple and Efficient Design for Semantic Segmentation with Transformers; 2021.
22 Sourget T, Hasany SN, Meriaudeau F, Petitjean C. Can SegFormer be a True Competitor to U-Net for Medical Image Segmentation? In: Waiter G, Lambrou T, Leontidis G, Oren N, Morris T, Gordon S, eds. Medical Image Understanding and Analysis. Vol. 14122. Cham: Springer Nature Switzerland; 2024:111-118.
23 Tizhoosh HR, Pantanowitz L. Artificial Intelligence and Digital Pathology: Challenges and Opportunities. J Pathol Inform. 2018;9:38. doi:10.4103/jpi.jpi_53_18.
24 Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B. Histopathological Image Analysis: A Review. IEEE Rev. Biomed. Eng. 2009;2:147-171. doi:10.1109/RBME.2009.2034865.
25 Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803-820. doi:10.1007/s00401-016-1545-1.
26 Le Hou, Samaras D, Kurc TM, Gao Y, Davis JE, Saltz JH. Patch-based Convolutional Neural Network for Whole Slide Tissue Image Classification. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit. 2016;2016:2424-2433. doi:10.1109/CVPR.2016.266.
27 Kumar N, Gupta R, Gupta S. Whole Slide Imaging (WSI) in Pathology: Current Perspectives and Future Directions. J Digit Imaging. 2020;33(4):1034-1040. doi:10.1007/s10278-020-00351-z.
28 Huang B, Reichman D, Collins LM, Bradbury K, Malof JM. Tiling and Stitching Segmentation Output for Remote Sensing: Basic Challenges and Recommendations; 2018.
29 Kolesnikov A, Beyer L, Zhai X, et al. Big Transfer (BiT): General Visual Representation Learning; 2019.
30 Maaz M, Shaker A, Cholakkal H, et al. EdgeNeXt: Efficiently Amalgamated CNN-Transformer Architecture for Mobile Vision Applications; 2022.
31 Simonyan K, Zisserman A. Very Deep Convolutional Networks for Large-Scale Image Recognition; 2014.
32 Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation; 2015.